Mosaic Level Image Processing¶
- Class:
- Alias:
mosaic_pipeline
The MosaicPipeline applies corrections to an overlapping group of images
and is setup to process only imaging observations.
This pipeline is used to apply the flux scale factor, flux,
determine a common background, skymatch, detect pixels the are
not consistent with the other datasets, outlier_detection, resample the image to a
single undistorted image, resample, and to run source_catalog
on the resulting corrected image.
The list of steps applied by the MosaicPipeline pipeline is shown in the
table below.
Step |
WFI-Image |
WFI-Prism |
WFI-Grism |
|---|---|---|---|
✓ |
𝕏 |
𝕏 |
|
✓ |
𝕏 |
𝕏 |
|
✓ |
𝕏 |
𝕏 |
|
✓ |
𝕏 |
𝕏 |
|
✓ |
𝕏 |
𝕏 |
Arguments¶
For more details about step arguments (including datatypes, possible values
and defaults) see romancal.pipeline.MosaicPipeline.spec.
--on_diskWhen
Truethe input association will be opened in a way that uses temporary files to avoid keeping all input models in memory.--resample_on_skycellFor outlier detection and resampling if the input association contains skycell information use it to compute the wcs to use for resampling.
You can see the options for strun using:
strun –help roman_mos
and this will list all the strun options as well as the step options for the roman_mos.
Inputs¶
An association of 2D calibrated image data¶
- Data model:
WfiImage- File suffix:
_cal
The input to the MosaicPipeline is a group of calibrated exposures,
e.g. “r0008308002010007027_0019_wfi01_cal.asdf”, which contains the
calibrated data for the the exposures. The most convenient way to pass the list of
exposures to be processed with the mosaic level pipeline is to use an association.
Instructions on how to create an input association an be found at asn_from_list.
The mosaic pipeline can create different types of products. In one mode you give it a list of calibrated images and the pipeline will run the above steps and the final product is a mosaic of the input images resampled to a regular grid. The mode is selected based on the target field in the association header. If the input association contains a target field which matches a skycell name (see TBD) then the mosaic pipeline will resample the final product onto the skycell grid.
If the association has been generated with skycell_asn the skycell name and projection coordinates should be available in the association header. If the skycell name is available and corresponds to a valid name in the database and the projection cell coordinates are not available in the association header then the pipeline will read the needed information from the data file containing the skycell information.
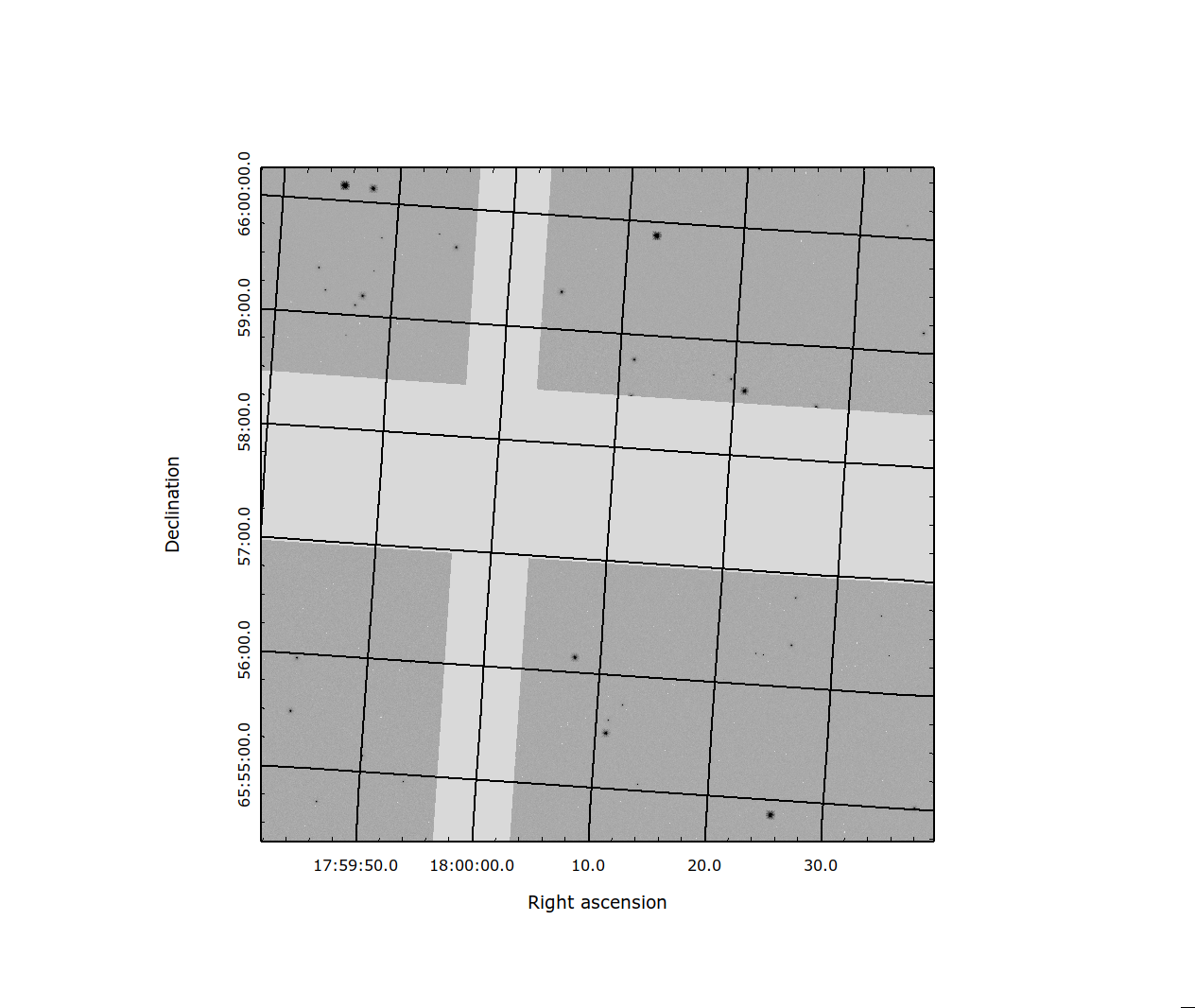
Fig. 1 Figure 1: Image showing the four SCA’s that overlap a given skycell.¶
The projection of the single WFI exposure resampled to a skycell is shown in Fig. 1. The image has the portion of the four SCAs that overlap the given skycell resampled to a regular grid on the WCS of the skycell. The gaps between the images show the placement of the SCAs in the detector. In general these gaps will be filled in by additional exposures in the visit.
If the target field does not contain a valid skycell name then the image or images will be resampled to a regular grid. To resample a single image the input will need to be an association with a single member.
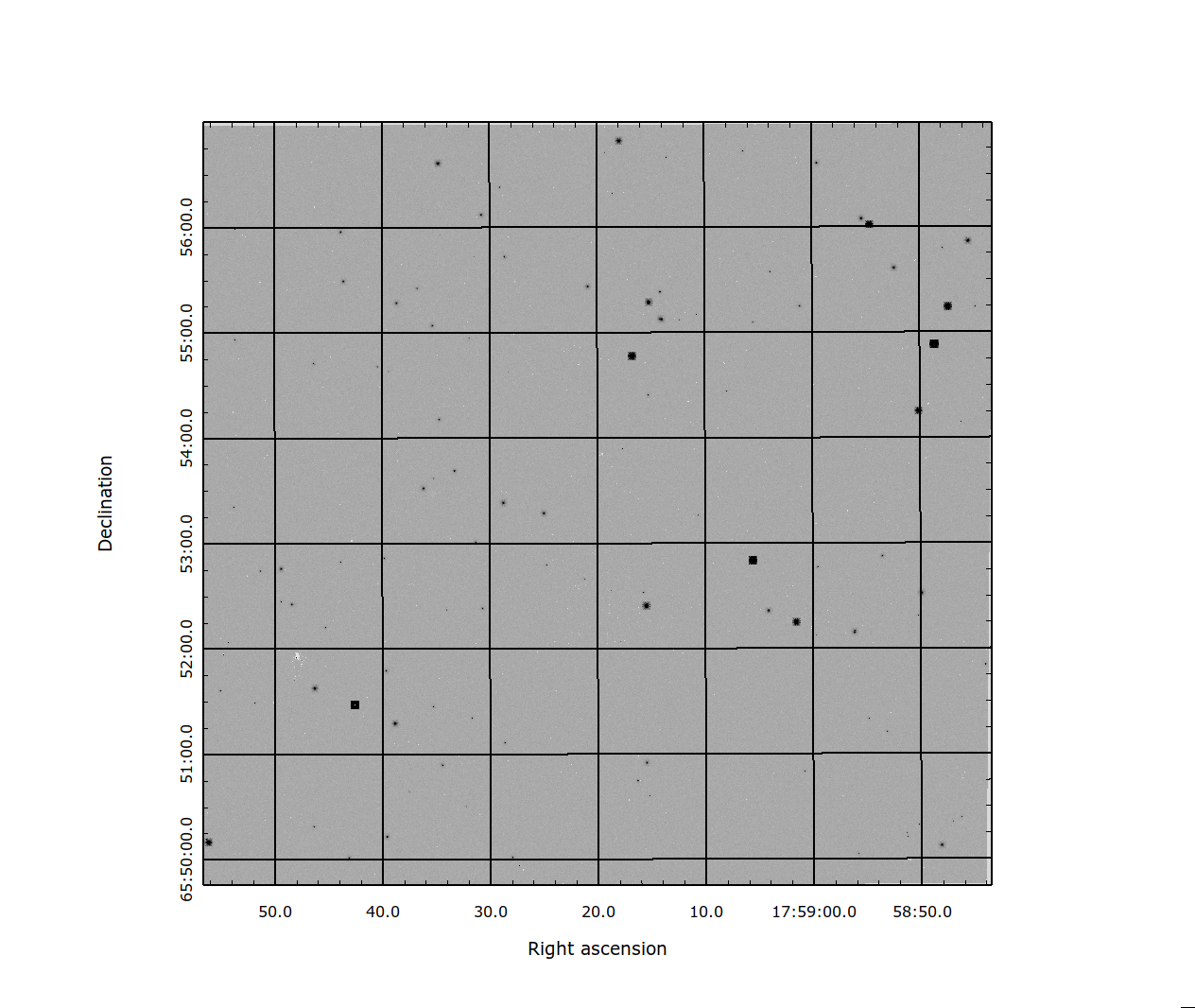
Fig. 2 Figure 2: An SCA resampled to a regular grid.¶ |
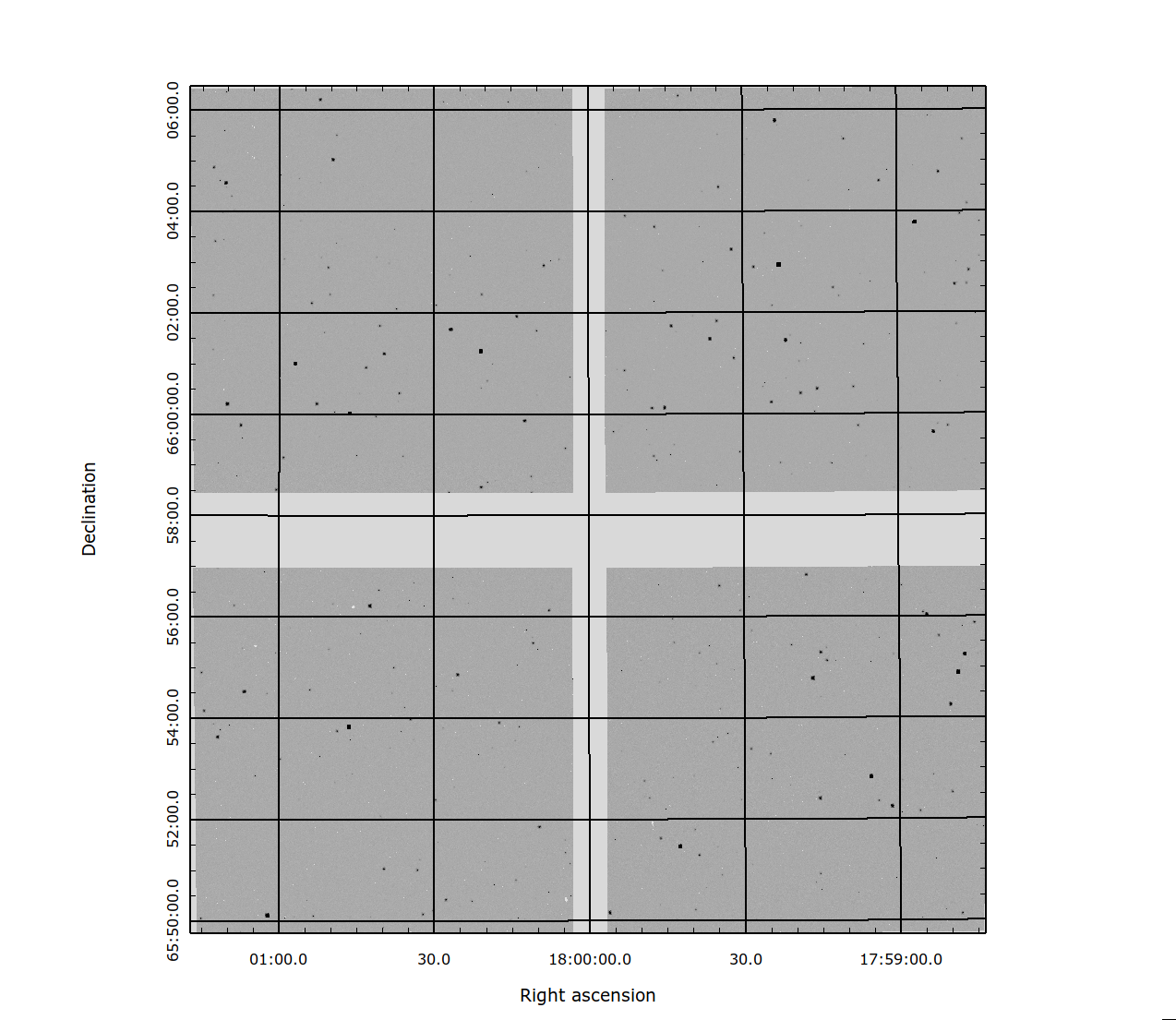
Fig. 3 Figure 3: Four SCAs resampled to a regular grid.¶ |
Figures 2 & 3 show the results of the mosaic pipeline on a single SCA and on four SCA’s in the WFI array. Using the code to mosaic large areas of the sky may result on the code needing large amounts of memory, so care is needed not to exceed your local memory limits when constructing mosaics in this manner.
Outputs¶
2D Image (MosaicModel)¶
The resampled data can be found in
- Data model:
WfiMosaic- File suffix:
_coadd
Catalog file (MosaicSourceCatalog)¶
The catalog data is in
- Data model:
MosaicSourceCatalog- File suffix:
_cat
Segmentation Map (SegmentationMapModel)¶
The segmentation map is
- Data model:
MosaicSegmentationMapModel- File suffix:
_segm
Result of applying all the mosaic level pipeline steps up through the source_catalog step is to produce data background corrected and cleaned of outliers and resampled to a distortion free grid along with the source catalog and segmentation map. The coadd file is 2D image data, with additional attributes for the mosaicing information. The cat file is an asdf file with the detected sources and the segmenation map is an asdf file linking the input images to the detected sources.